RandomWalkwithRestart.jar:
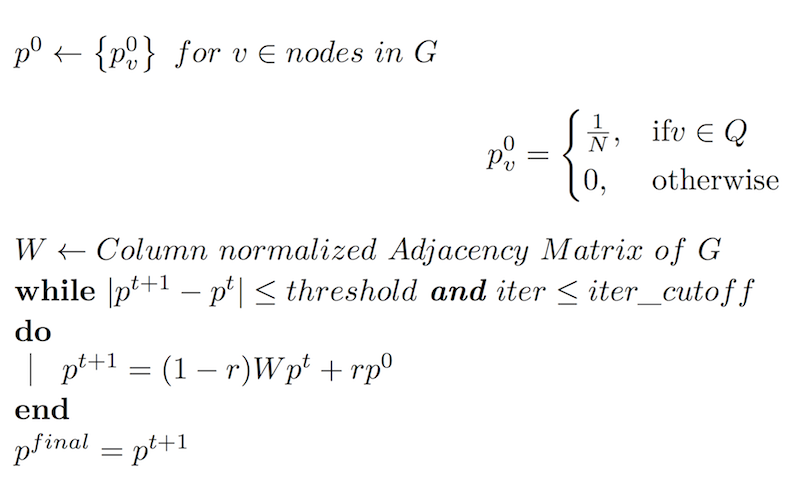
Let G = (V, E) be the graph representing a pairwise sequence similarity network, where V is the set of nodes (sequences), and E is the set of weighted undirected edges, where the bit-score from BLAST weight, which shows the strength of similarity between sequence pairs, is assigned as edge weights. We define the homology of a node v to set of start nodes Q, pq(v), as follows:
The random walk with restart method simulates a random walker that starts from one of the start nodes in q. At every step, the walker selects among the available neighbors, based on edge weights of the neighbors of the node he is currently in, with probability (1-r), or goes back to query set Q, with probability r. The restart probability r imposes a restriction on how far the random walker can get away from the start nodes Q. With r close to 1, the probability vector reflects the local structure around Q, and as r gets close to 0, a more global view is observed.
The local structure around Q being the determinant for identifying sequence homology, the default setting for r in Orthofuzz is 0.33.
The probability pvt, the probability of the random walker to be at node v at step t, is initialized to 1/N for nodes in the query set, where N is the number of nodes in the query set and to 0 for all the remaining nodes.
The steady state probability pvfinal is a measure of the homology of the node final to the query set Q. The p final is estimated using iterative matrix operations shown in figure 1.